 Utility functions for computing the expectation, variance and standard
deviation of the distributions are also available. These functions use
numerical integration.
Utility functions for computing the expectation, variance and standard
deviation of the distributions are also available. These functions use
numerical integration.The ibdsegments package offers functions related to identity by descent (IBD) probability for pedigree members. Both the discrete case (identity states) and the continuous case (segment lengths) are treated using a Hidden Markov Model (HMM) approach. Key functionality includes:
The main advantage of this approach is the flexibility to define complex IBD states such as IBD among more than two pedigree members. However, it is limited to relatively small pedigrees, as the state space grows exponentially with the number of non-founders in the pedigree.
The ibdsegments package is still in development and
not yet available from CRAN. You can install the
ibdsegments package using the pak package in R:
# install.packages("pak")
pak::pak("mkruijver/ibdsegments")library(ibdsegments)
#>
#> Attaching package: 'ibdsegments'
#> The following objects are masked from 'package:stats':
#>
#> sd, varThe d_ibd function may be used to compute identity
coefficients. The example shows how to compute the kappa
coefficients for half siblings.
ped_hs <- pedtools::halfSibPed()
d_ibd(0, pedigree = ped_hs, states = "kappa")
#> [1] 0.5
d_ibd(1, pedigree = ped_hs, states = "kappa")
#> [1] 0.5Besides kappa, other identity states supported
include:
ibd: 0, 1, or 2 whenever all selected pedigree members
jointly share this number of founder allele labelsidentity: Jacquard’s 9 condensed identity statesdetailed: the 15 detailed identity statesFor example, the inbreeding coefficient may be computed as an
ibd state for a single pedigree member.
ped_inbred <- pedtools::fullSibMating(n = 1)
d_ibd(ibd = 1, pedigree = ped_inbred, ids = 5, states = "ibd")
#> [1] 0.25The probability that three siblings are jointly double ibd is also
easily computed using the d_ibd function.
ped_3fs <- pedtools::nuclearPed(nch = 3)
d_ibd(ibd = 2, pedigree = ped_3fs, states = "ibd")
#> [1] 0.0625The identity coefficients computed above are IBD probabilities at
single positions on a chromosome. Taking a continuous view, the fraction
of the chromosome that is in each of the states is in expectation equal
to these IBD probabilities. The r_cibd functions implements
random sampling of continuous IBD:
set.seed(1)
r_cibd(n = 1, pedigree = ped_hs, states = "kappa", chromosome_length = 100)
#> $samples
#> sample chromosome start end length state
#> 1 1 1 0.00000 59.08214 59.082139 1
#> 2 1 1 59.08214 66.07190 6.989763 0
#> 3 1 1 66.07190 82.15168 16.079779 1
#> 4 1 1 82.15168 100.00000 17.848319 0
#>
#> $stats
#> total_length segment_count
#> 1 75.16192 2The total_ibd_dist function obtains the full
distribution of the total length of IBD segments across a chromosome.
For example, we may obtain this distribution for half siblings on a
chromosome with a length of 100 cM.
d_hs <- total_ibd_dist(ped_hs, chromosome_length = 100)
d_hs
#> Probability distribution of total length of segments in ibd state 1
#> Chromosome length: 100 cM
#>
#> Weight of continuous density: 0.8646647
#>
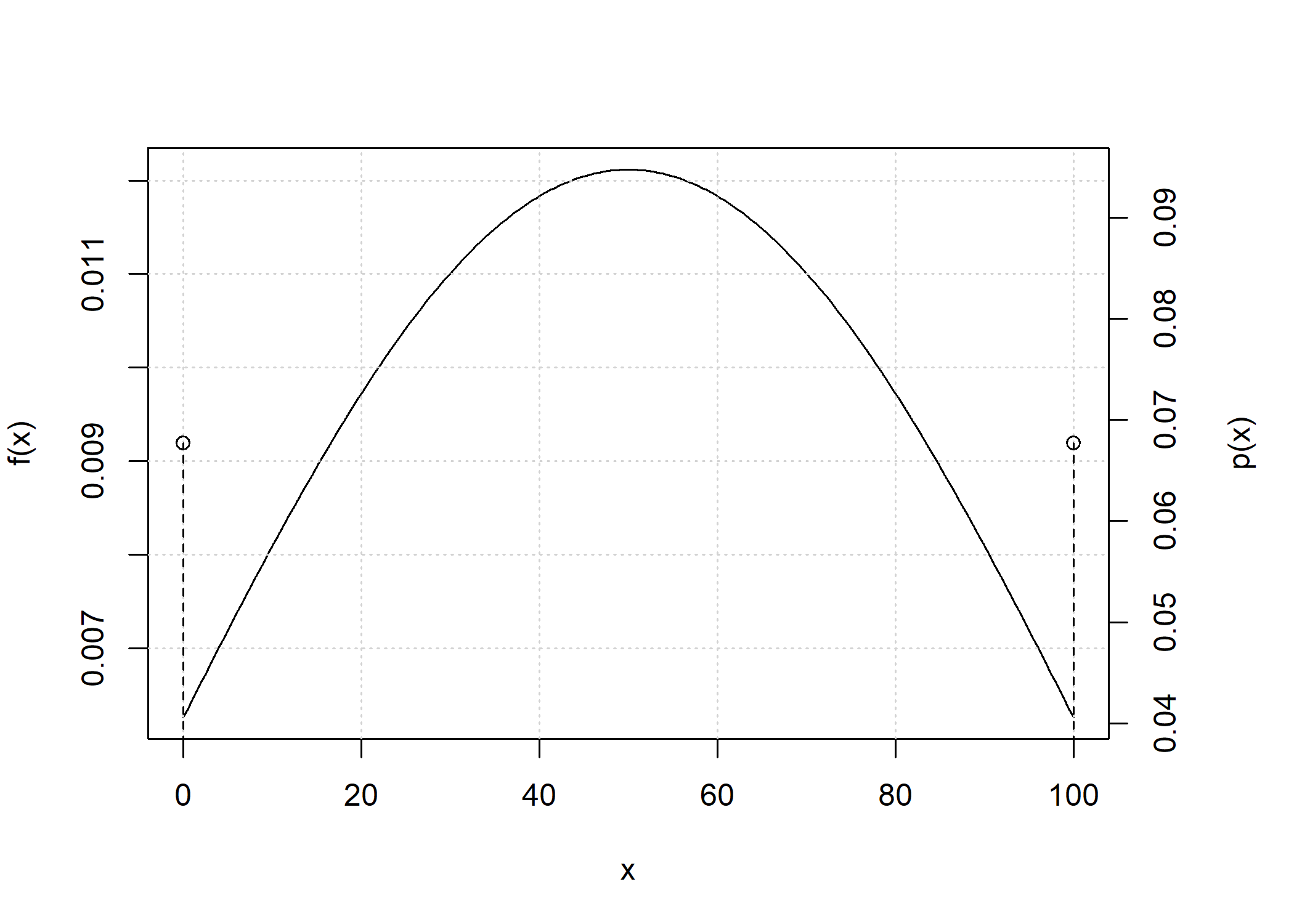
#> Point masses:
#> x px
#> 0 0.06766764
#> 100 0.06766764The distribution has two point masses (no IBD at all and fully IBD) and admits a density function otherwise. A plot includes both components.
plot(d_hs) Utility functions for computing the expectation, variance and standard
deviation of the distributions are also available. These functions use
numerical integration.
Utility functions for computing the expectation, variance and standard
deviation of the distributions are also available. These functions use
numerical integration.
E(d_hs)
#> [1] 50
sd(d_hs)
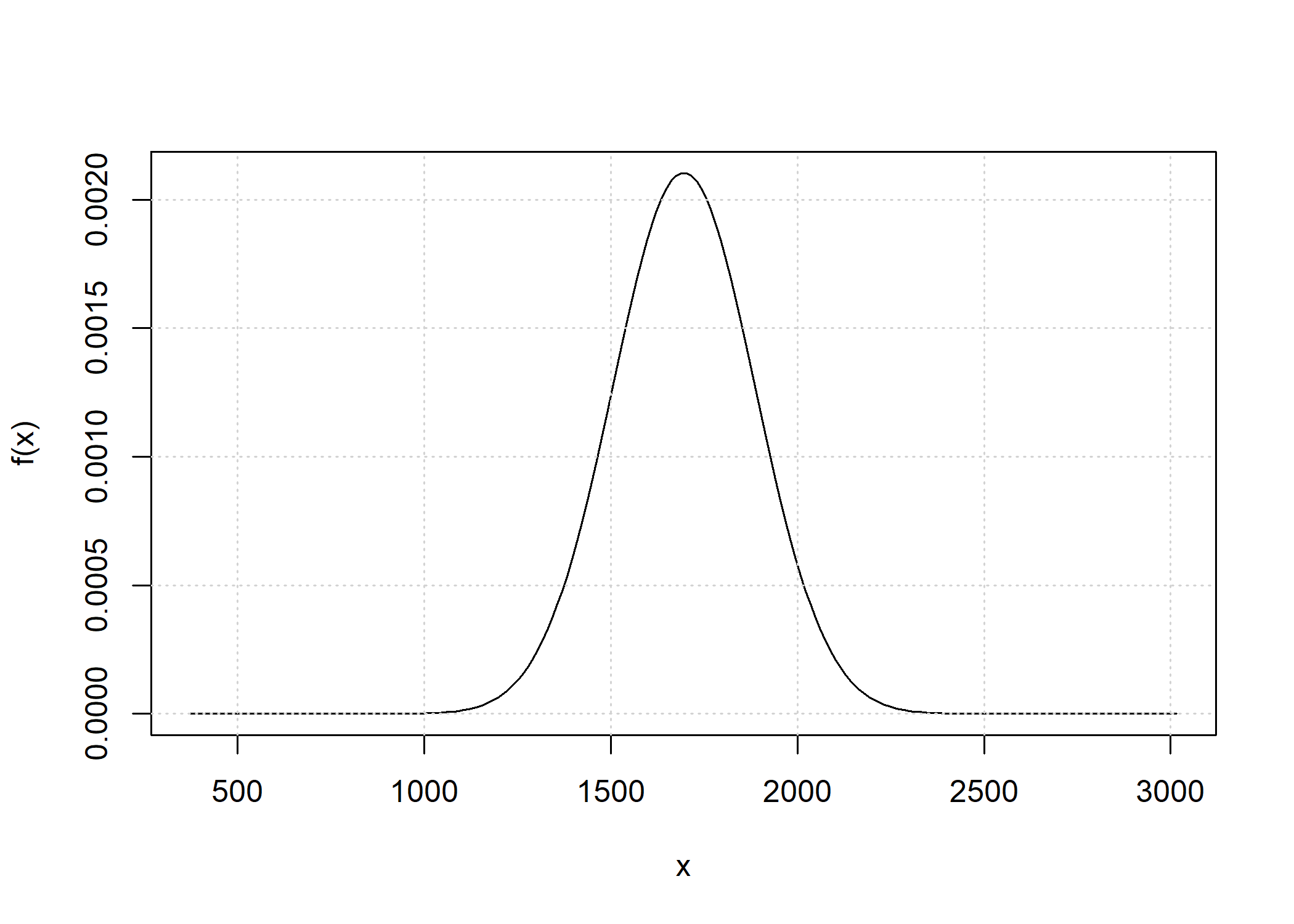
#> [1] 30.71195The convolution of the total IBD distribution across chromosomes is
obtained when the chromosome_length parameter has length
greater than 1.
d_hs_conv <- total_ibd_dist(ped_hs,
chromosome_length = c(250, 200, 150, 150, 100))
plot(d_hs_conv)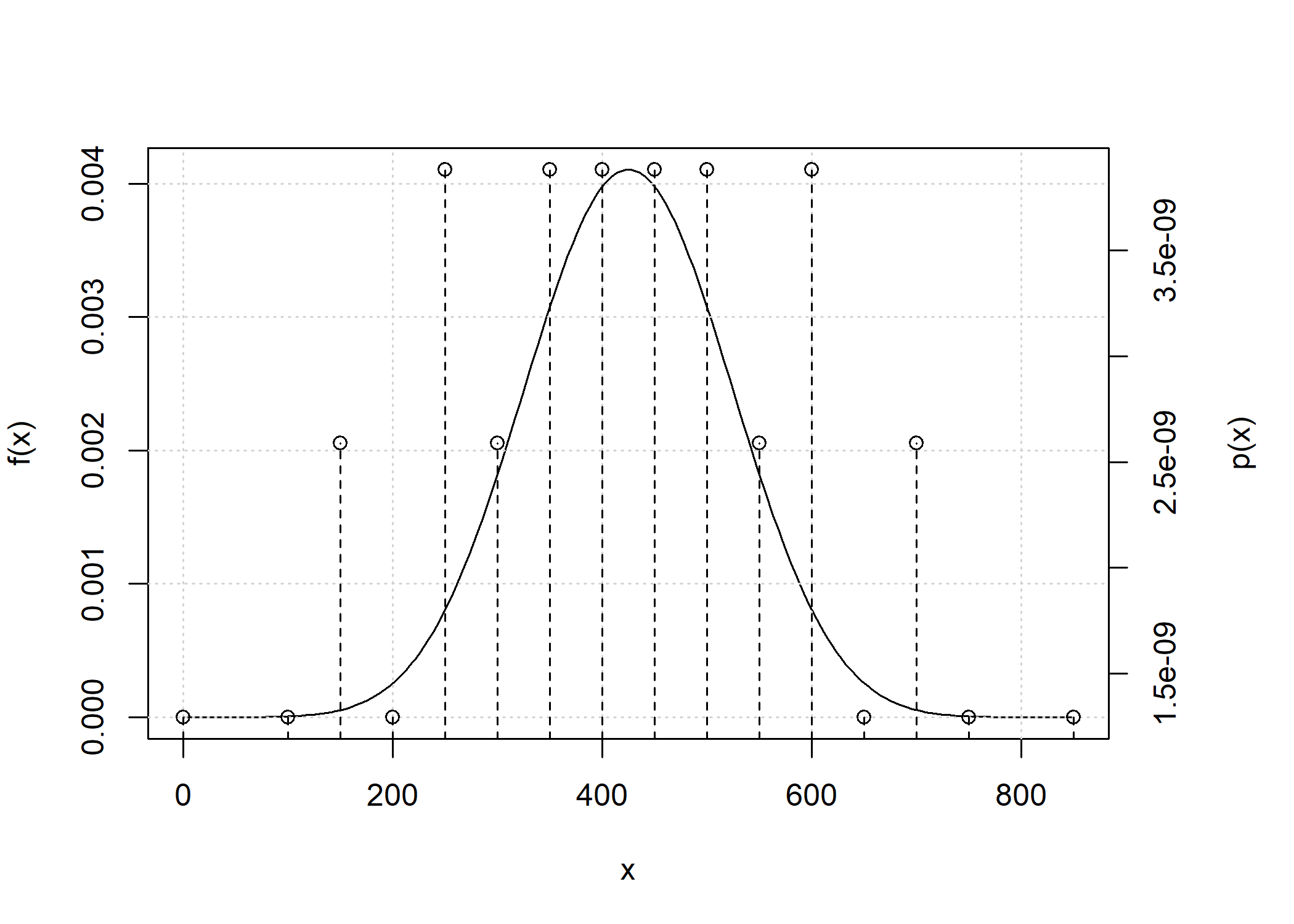
Because the number of point masses may increase quickly, by default
any point mass below 1e-9 is removed.
L <- c(267.77, 251.73, 218.31, 202.89, 197.08, 186.02, 178.4, 161.54,
157.35, 169.28, 154.5, 165.49, 127.23, 116, 117.32, 126.59, 129.53,
116.52, 106.35, 107.76, 62.88, 70.84)
d_hs_full_conv <- total_ibd_dist(ped_hs, chromosome_length = L)
d_hs_full_conv
#> Probability distribution of total length of segments in ibd state 1
#> Chromosome length: 62.88 70.84 106.35 107.76 116 116.52 117.32 126.59 127.23 129.53 154.5 157.35 161.54 165.49 169.28 178.4 186.02 197.08 202.89 218.31 251.73 267.77 cM
#>
#> Weight of continuous density: 1
#>
#> Point masses:
#> [1] x px
#> <0 rows> (or 0-length row.names)
plot(d_hs_full_conv)